Asthma and heart disease in NY state
- Considering geological factor as an important predictor, we calculated percentage of asthma cases based on their locations with respect to counties in NY state and created maps with their asthma rate based on colors among counties in NY state.If the color is grey, that means there was no respondents in those counties at that year.
Asthma Map
According to the graphs, we can know that:
- In 2003,
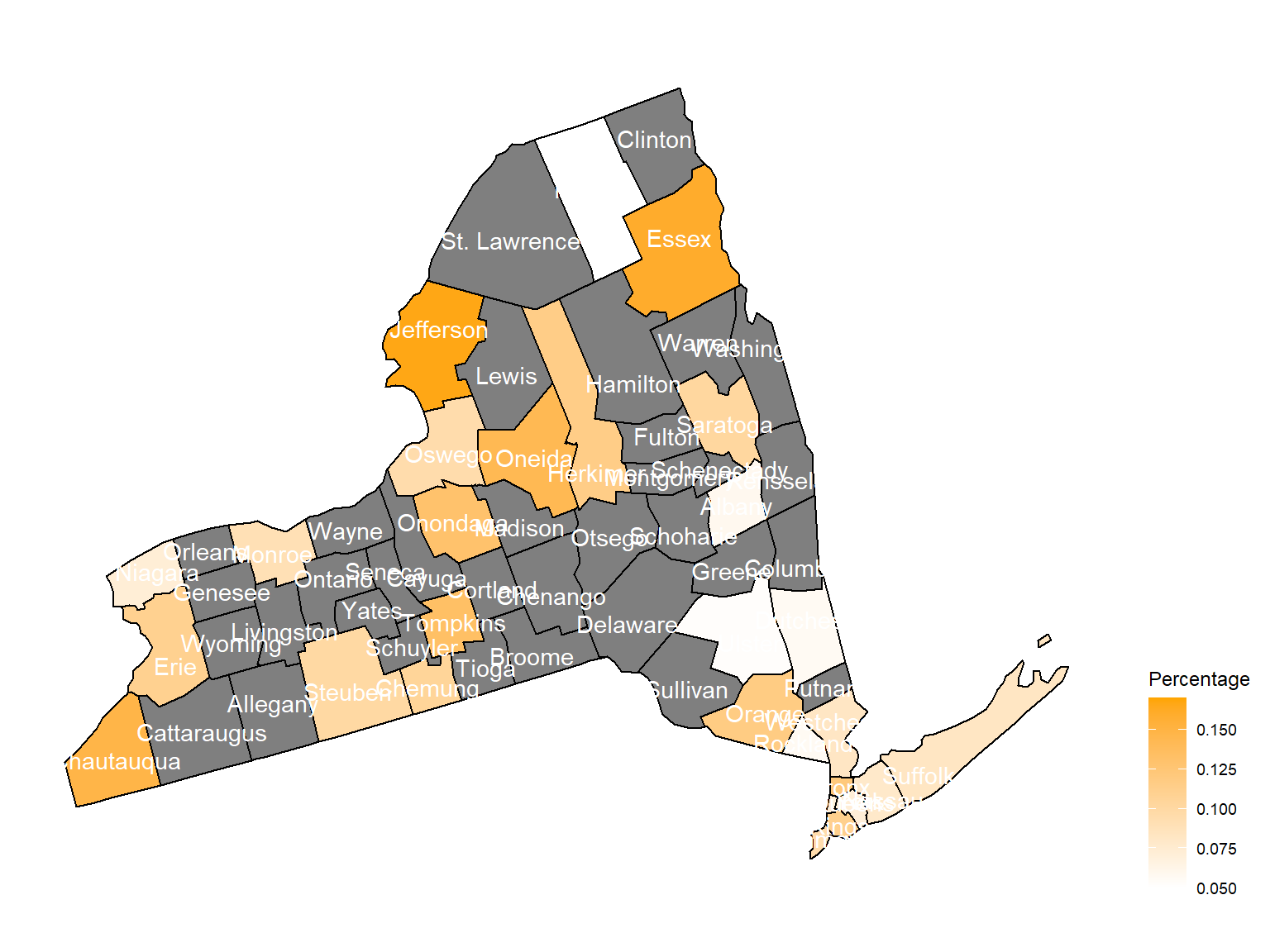
Chautauquacounty is with the most asthma patients. After that, the percentage decreased until 2009. - After 2008, asthma patients in
SteubenandChemungwere increasing. - Counties in north of New York, including
Jefferson,St. Lawrence,FranklinandEssexare also with higher percentage of asthma patients. - Urban counties, including
Bronx,QueensandErie, are also with a higher percentage of asthma patients.
2003
asthma_now_county_df1 =
brfss_air_df %>%
mutate(
fips = str_c(state_code.x,county_code)
) %>%
group_by(state_code.x, county_code,county,fips, year) %>%
filter(year == 2003) %>%
count(
county,asthma_status
) %>%
mutate(
percent = n/sum(n)
) %>%
filter(asthma_status == "1") %>%
spread(asthma_status, percent)
asthma_now_county_plot_map =
plot_usmap(regions = "county", include = c("NY"), data = asthma_now_county_df1, values = "1", label = TRUE, label_color = "White") +
scale_fill_continuous(
low = "white", high = "Orange", name = "Percentage", label = scales::comma, limits = c(0.05,0.17)
) +
theme(legend.position = "right")
asthma_now_county_plot_map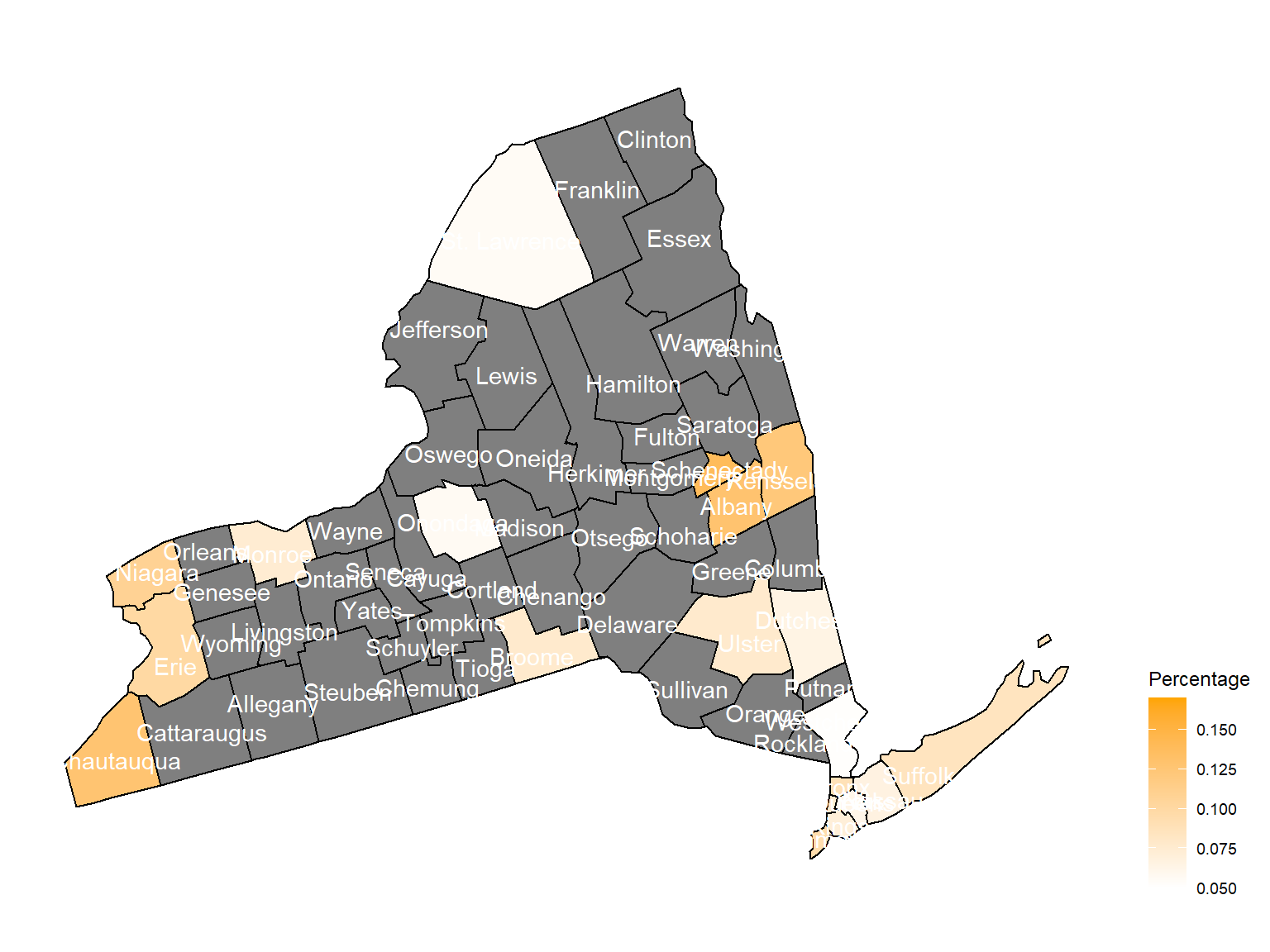
2004
asthma_now_county_df2 =
brfss_air_df %>%
mutate(
fips = str_c(state_code.x,county_code)
) %>%
group_by(state_code.x, county_code,county,fips, year) %>%
filter(year == 2004) %>%
count(
county,asthma_status
) %>%
mutate(
percent = n/sum(n)
) %>%
filter(asthma_status == "1") %>%
spread(asthma_status, percent)
asthma_now_county_plot_map =
plot_usmap(regions = "county", include = c("NY"), data = asthma_now_county_df2, values = "1", label = TRUE, label_color = "White") +
scale_fill_continuous(
low = "white", high = "Orange", name = "Percentage", label = scales::comma, limits = c(0.05,0.17)
) +
theme(legend.position = "right")
asthma_now_county_plot_map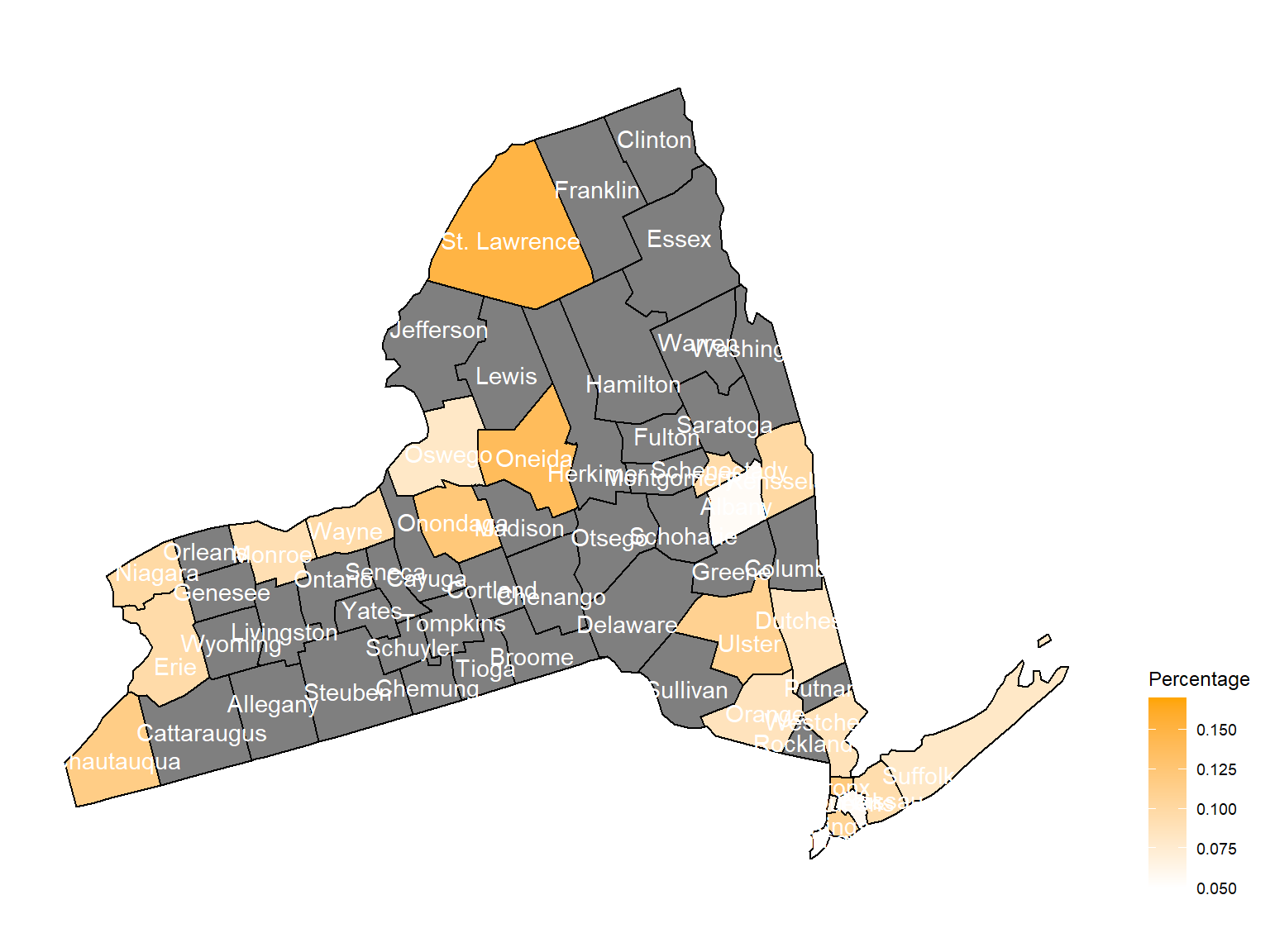
2005
asthma_now_county_df3 =
brfss_air_df %>%
mutate(
fips = str_c(state_code.x,county_code)
) %>%
group_by(state_code.x, county_code,county,fips, year) %>%
filter(year == 2005) %>%
count(
county,asthma_status
) %>%
mutate(
percent = n/sum(n)
) %>%
filter(asthma_status == "1") %>%
spread(asthma_status, percent)
asthma_now_county_plot_map =
plot_usmap(regions = "county", include = c("NY"), data = asthma_now_county_df3, values = "1", label = TRUE, label_color = "White") +
scale_fill_continuous(
low = "white", high = "Orange", name = "Percentage", label = scales::comma, limits = c(0.05,0.17)
) +
theme(legend.position = "right")
asthma_now_county_plot_map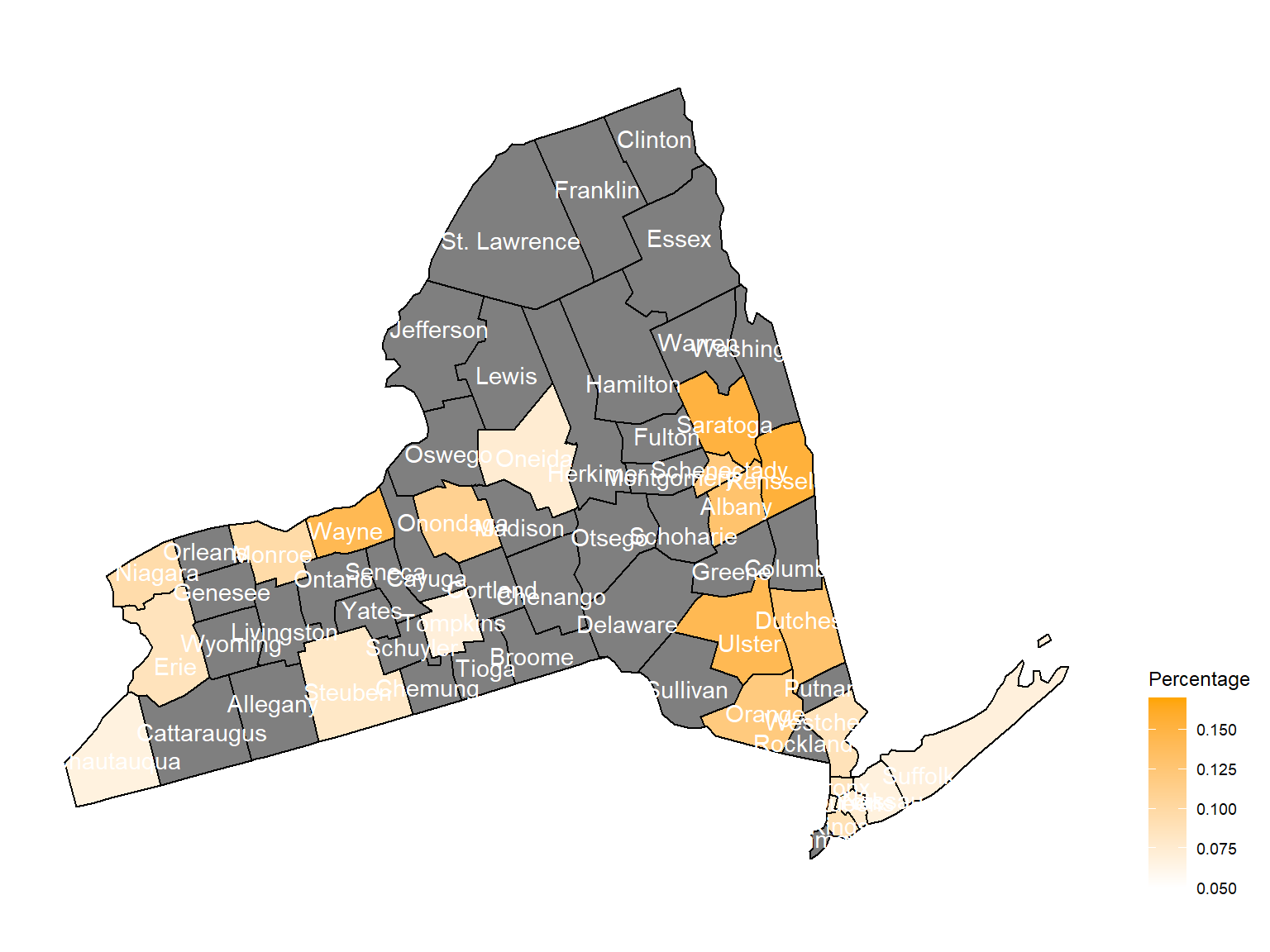
2006
asthma_now_county_df4 =
brfss_air_df %>%
mutate(
fips = str_c(state_code.x,county_code)
) %>%
group_by(state_code.x, county_code,county,fips, year) %>%
filter(year == 2006) %>%
count(
county,asthma_status
) %>%
mutate(
percent = n/sum(n)
) %>%
filter(asthma_status == "1") %>%
spread(asthma_status, percent)
asthma_now_county_plot_map =
plot_usmap(regions = "county", include = c("NY"), data = asthma_now_county_df4, values = "1", label = TRUE, label_color = "White") +
scale_fill_continuous(
low = "white", high = "Orange", name = "Percentage", label = scales::comma, limits = c(0.05,0.17)
) +
theme(legend.position = "right")
asthma_now_county_plot_map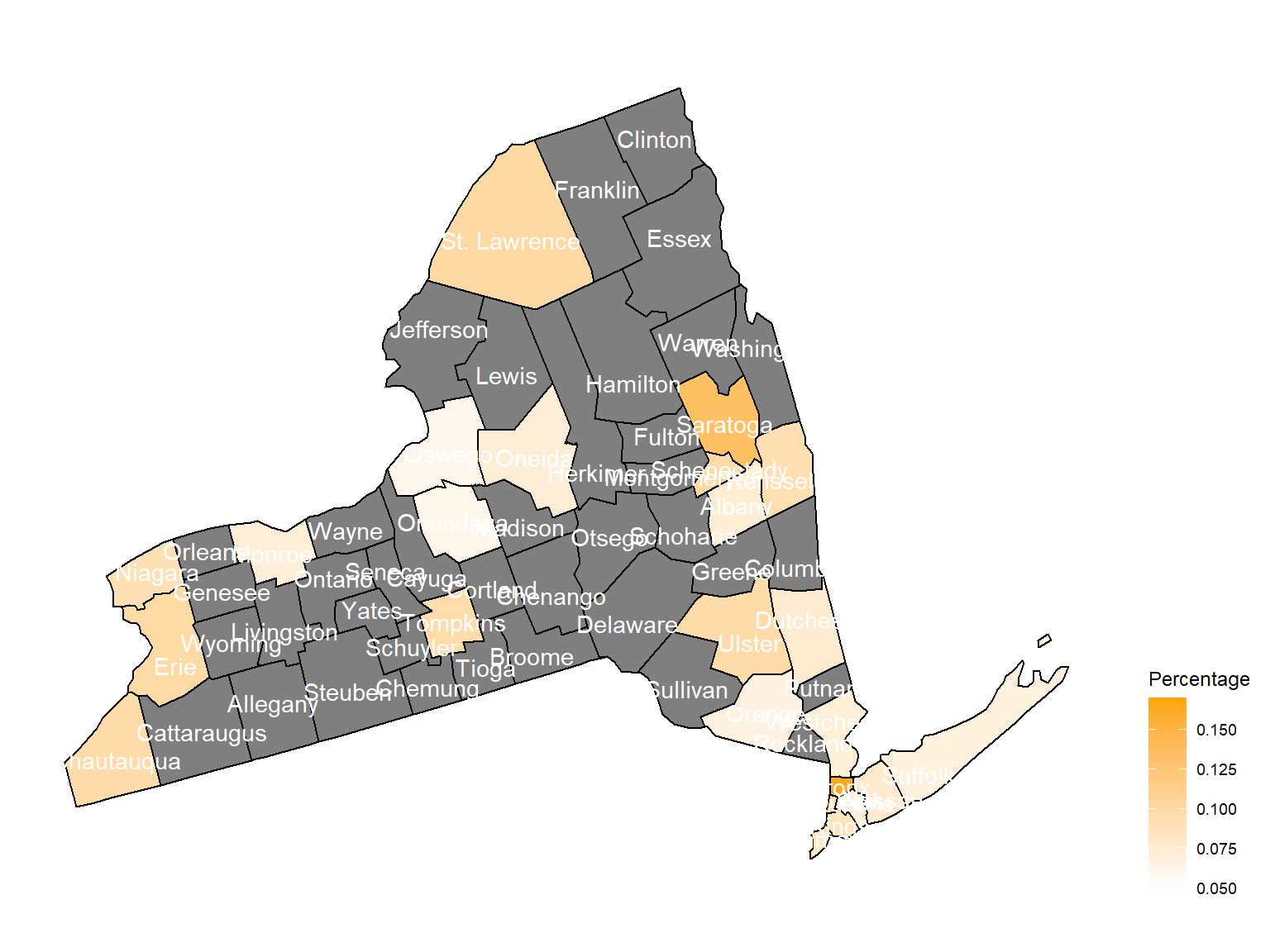
2007
asthma_now_county_df5 =
brfss_air_df %>%
mutate(
fips = str_c(state_code.x,county_code)
) %>%
group_by(state_code.x, county_code,county,fips, year) %>%
filter(year == 2007) %>%
count(
county,asthma_status
) %>%
mutate(
percent = n/sum(n)
) %>%
filter(asthma_status == "1") %>%
spread(asthma_status, percent)
asthma_now_county_plot_map =
plot_usmap(regions = "county", include = c("NY"), data = asthma_now_county_df5, values = "1", label = TRUE, label_color = "White") +
scale_fill_continuous(
low = "white", high = "Orange", name = "Percentage", label = scales::comma, limits = c(0.05,0.17)
) +
theme(legend.position = "right")
asthma_now_county_plot_map
2008
asthma_now_county_df6 =
brfss_air_df %>%
mutate(
fips = str_c(state_code.x,county_code)
) %>%
group_by(state_code.x, county_code,county,fips, year) %>%
filter(year == 2008) %>%
count(
county,asthma_status
) %>%
mutate(
percent = n/sum(n)
) %>%
filter(asthma_status == "1") %>%
spread(asthma_status, percent)
asthma_now_county_plot_map =
plot_usmap(regions = "county", include = c("NY"), data = asthma_now_county_df6, values = "1", label = TRUE, label_color = "White") +
scale_fill_continuous(
low = "white", high = "Orange", name = "Percentage", label = scales::comma, limits = c(0.05,0.17)
) +
theme(legend.position = "right")
asthma_now_county_plot_map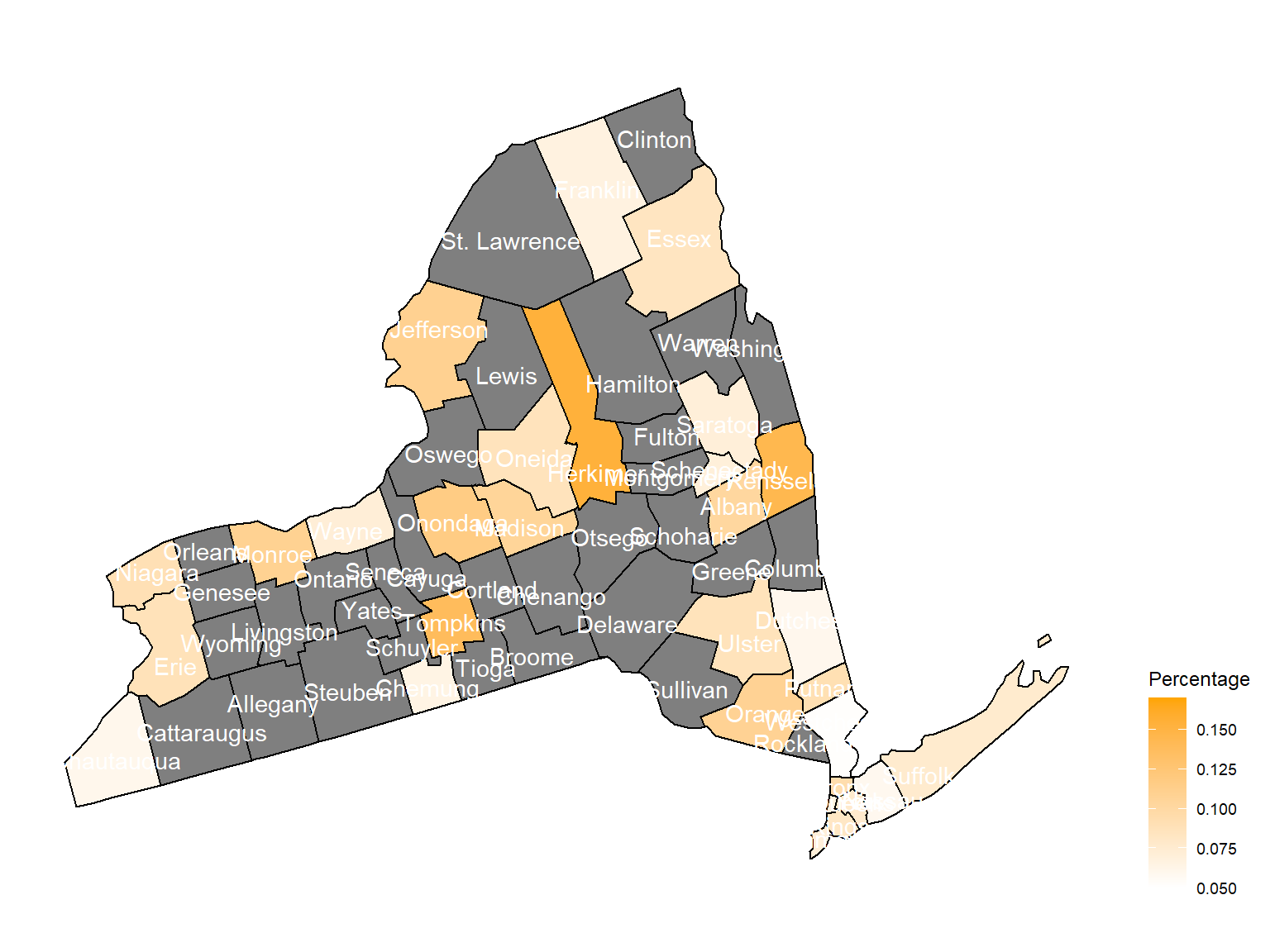
2009
asthma_now_county_df7 =
brfss_air_df %>%
mutate(
fips = str_c(state_code.x,county_code)
) %>%
group_by(state_code.x, county_code,county,fips, year) %>%
filter(year == 2009) %>%
count(
county,asthma_status
) %>%
mutate(
percent = n/sum(n)
) %>%
filter(asthma_status == "1") %>%
spread(asthma_status, percent)
asthma_now_county_plot_map =
plot_usmap(regions = "county", include = c("NY"), data = asthma_now_county_df7, values = "1", label = TRUE, label_color = "White") +
scale_fill_continuous(
low = "white", high = "Orange", name = "Percentage", label = scales::comma, limits = c(0.05,0.17)
) +
theme(legend.position = "right")
asthma_now_county_plot_map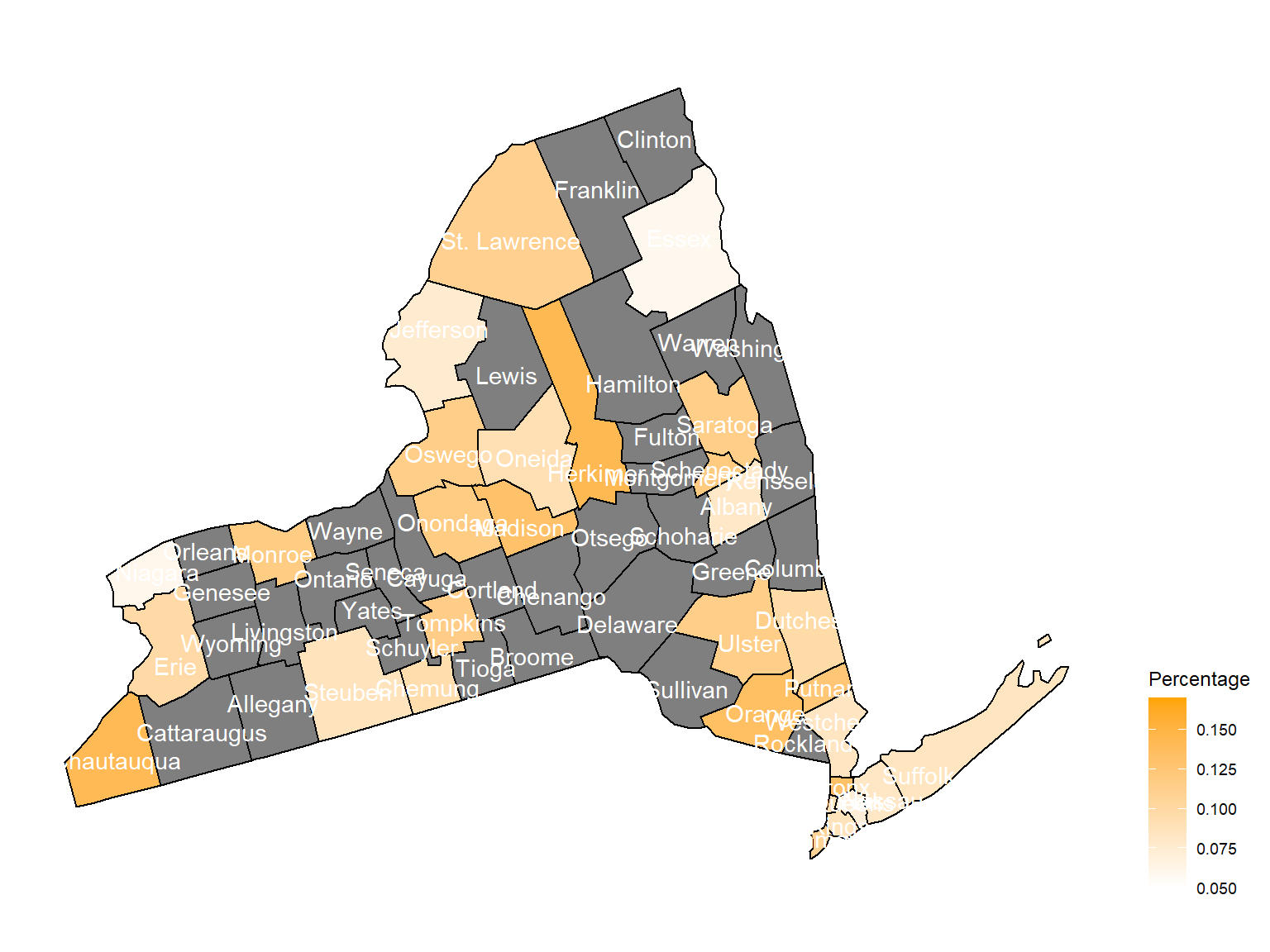
2010
asthma_now_county_df8 =
brfss_air_df %>%
mutate(
fips = str_c(state_code.x,county_code)
) %>%
group_by(state_code.x, county_code,county,fips, year) %>%
filter(year == 2010) %>%
count(
county,asthma_status
) %>%
mutate(
percent = n/sum(n)
) %>%
filter(asthma_status == "1") %>%
spread(asthma_status, percent)
asthma_now_county_plot_map =
plot_usmap(regions = "county", include = c("NY"), data = asthma_now_county_df8, values = "1", label = TRUE, label_color = "White") +
scale_fill_continuous(
low = "white", high = "Orange", name = "Percentage", label = scales::comma, limits = c(0.05,0.17)
) +
theme(legend.position = "right")
asthma_now_county_plot_map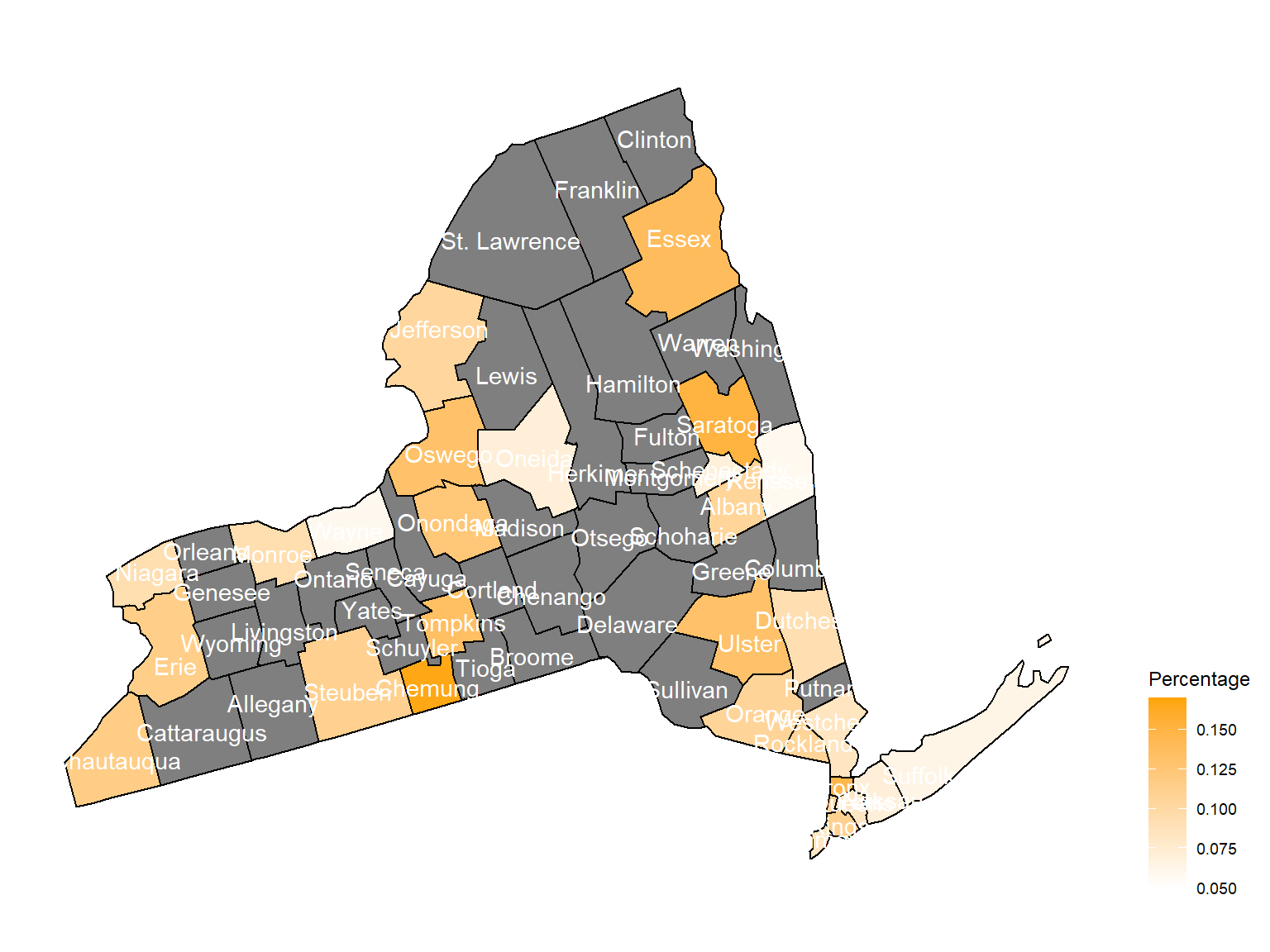
2011
asthma_now_county_df9 =
brfss_air_df %>%
mutate(
fips = str_c(state_code.x,county_code)
) %>%
group_by(state_code.x, county_code,county,fips, year) %>%
filter(year == 2011) %>%
count(
county,asthma_status
) %>%
mutate(
percent = n/sum(n)
) %>%
filter(asthma_status == "1") %>%
spread(asthma_status, percent)
asthma_now_county_plot_map =
plot_usmap(regions = "county", include = c("NY"), data = asthma_now_county_df9, values = "1", label = TRUE, label_color = "White") +
scale_fill_continuous(
low = "white", high = "Orange", name = "Percentage", label = scales::comma, limits = c(0.05,0.17)
) +
theme(legend.position = "right")
asthma_now_county_plot_map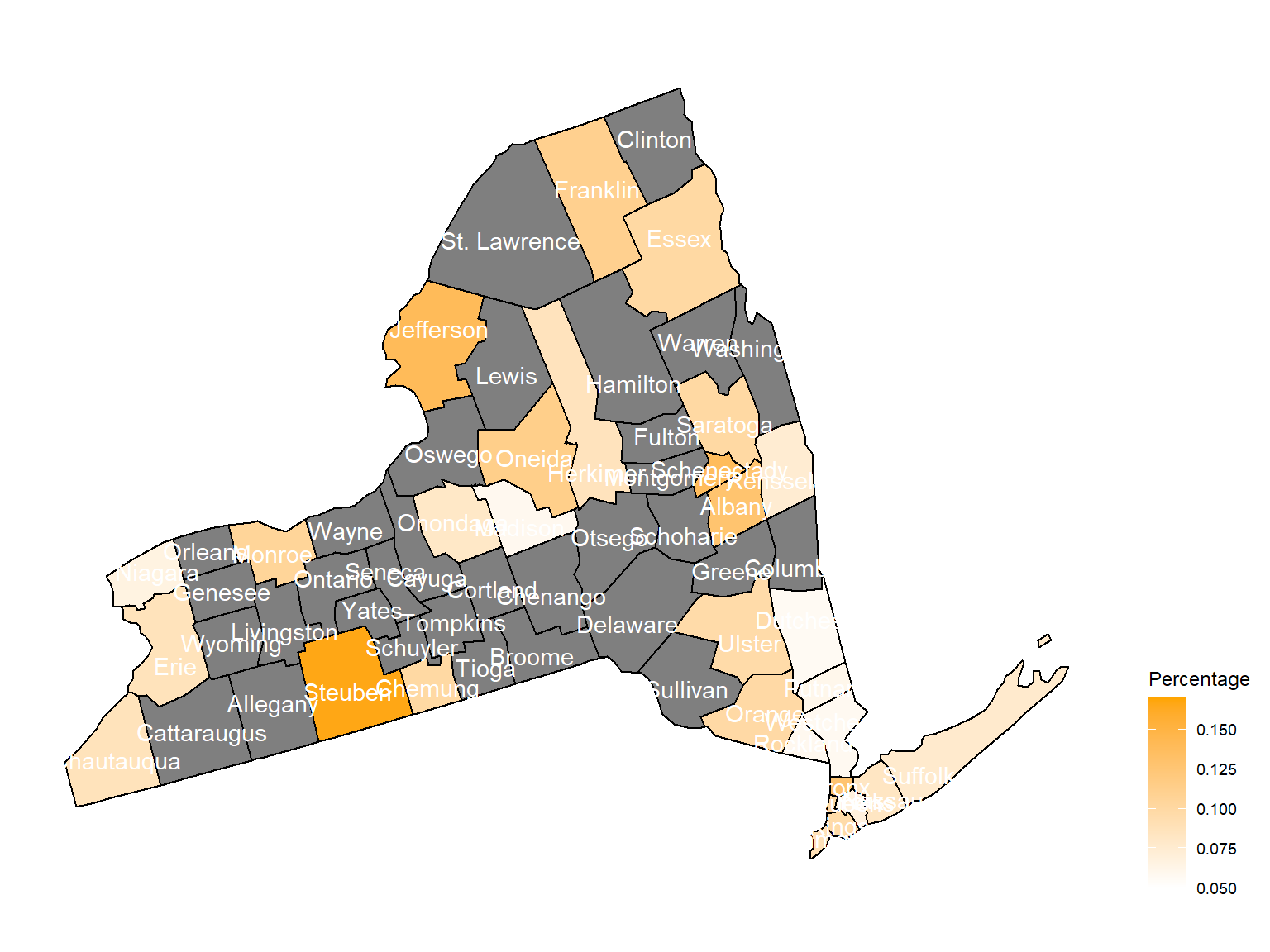
2012
asthma_now_county_df10 =
brfss_air_df %>%
mutate(
fips = str_c(state_code.x,county_code)
) %>%
group_by(state_code.x, county_code,county,fips, year) %>%
filter(year == 2012) %>%
count(
county,asthma_status
) %>%
mutate(
percent = n/sum(n)
) %>%
filter(asthma_status == "1") %>%
spread(asthma_status, percent)
asthma_now_county_plot_map =
plot_usmap(regions = "county", include = c("NY"), data = asthma_now_county_df10, values = "1", label = TRUE, label_color = "White") +
scale_fill_continuous(
low = "white", high = "Orange", name = "Percentage", label = scales::comma, limits = c(0.05,0.17)
) +
theme(legend.position = "right")
asthma_now_county_plot_map